Agricultural Applications
(CVPR'22) GrainSpace: A Large-scale Dataset for Fine-grained and Domain-adaptive Recognition of Cereal Grains
Lei Fan, Yiwen Ding, Dongdong Fan, Donglin Di, Maurice Pagnucco, Yang Song
Cereal grains are a vital part of human diets and are important commodities for people’s livelihood and international trade. Grain Appearance Inspection (GAI) serves as one of the crucial steps for the determination of grain quality and grain stratification for proper circulation, storage and food processing, etc.
In this paper we formulate GAI as three ubiquitous computer vision tasks: fine-grained recognition, domain adaptation and out-of-distribution recognition. We present a large-scale and publicly available cereal grains dataset called GrainSpace. Specifically, we construct three types of device prototypes for data acquisition, and a total of 5.25 million images determined by professional inspectors. The grain samples including wheat, maize and rice are collected from five countries and more than 30 regions.

Histopathology Image Analysis
(MICCAI'22) Fast FF-to-FFPE Whole Slide Image Translation via Laplacian Pyramid and Contrastive Learning
Lei Fan, Arcot Sowmya, Erik Meijering, Yang Song
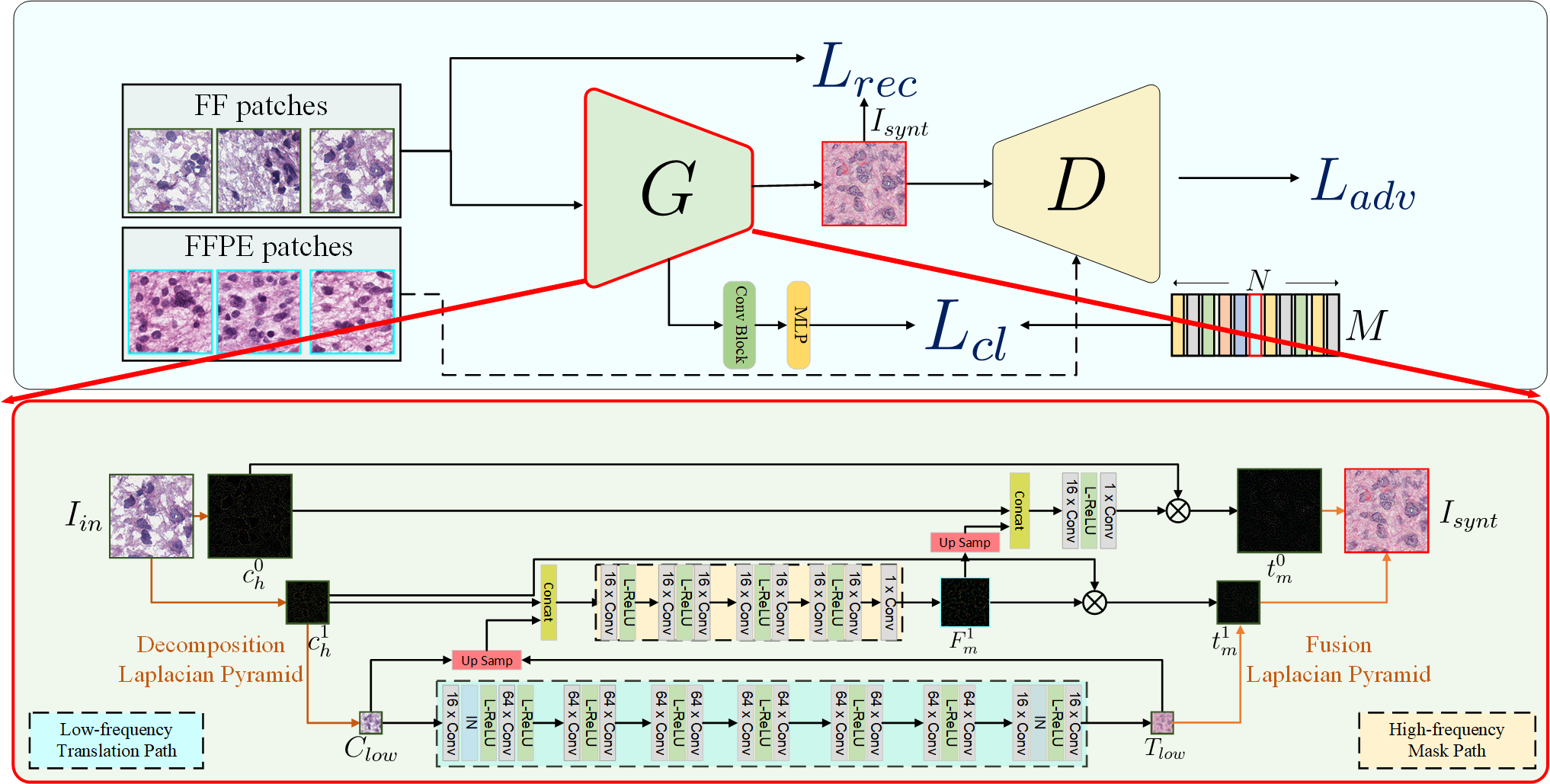
Formalin-Fixed Paraffin-Embedded (FFPE) and Fresh Frozen (FF) are two major types of histopathological Whole Slide Images (WSIs). FFPE provides high-quality images, however the acquisition process usually takes 12 to 48 h, while FF with relatively low-quality images takes less than 15 min to acquire. In this work, we focus on the task of translating FF to FFPE style (FF2FFPE), to synthesize FFPE-style images from FF images. However, WSIs with giga-pixels impose heavy constraints on computation and time resources.
To address these issues, we propose the fastFF2FFPE for translating FF into FFPE-style efficiently. Specifically, we decompose FF images into low- and high-frequency components based on the Laplacian Pyramid, wherein the low-frequency component at low resolution is transformed into FFPE-style with low computational cost, and the high-frequency component is used for providing details. We further employ contrastive learning to encourage similarities between original and output patches. We conduct FF2FFPE translation experiments on The Cancer Genome Atlas (TCGA) Glioblastoma Multiforme (GBM) and Lung Squamous Cell Carcinoma (LUSC) datasets, and verify the efficacy of our model on Microsatellite Instability prediction in gastrointestinal cancer.
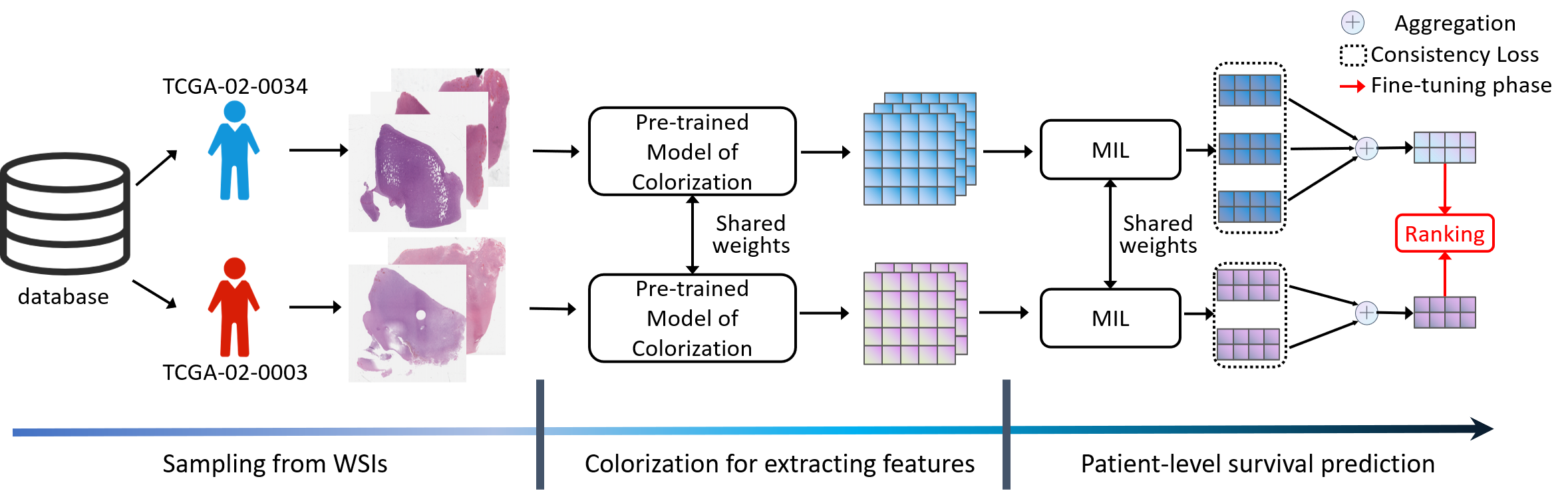
(MICCAI'21) Learning Visual Features by Colorization for Slide-Consistent Survival Prediction from Whole Slide Images (Student Travel Award)
Lei Fan, Arcot Sowmya, Erik Meijering, Yang Song
Recent deep learning techniques have shown promising performance on survival prediction from Whole Slide Images (WSIs). These methods are often based on multiple-step frameworks including patch sampling, feature extraction, and feature aggregation. However, feature extraction typically relies on handcrafted features or Convolutional Neural Networks (CNNs) pretrained on ImageNet without fine-tuning, thus leading to suboptimal performance. Besides, to aggregate features, previous studies focus on WSI-level survival prediction but ignore the heterogeneous information that is present in multiple WSIs acquired for the same patient.
To address the above challenges, we propose a survival prediction model that exploits heterogeneous features at the patient-level. Specifically, we introduce colorization as the pretext task to train the CNNs which are tailored for extracting features from patches of WSIs. In addition, we develop a patient-level framework integrating multiple WSIs for survival prediction with consistency and ranking losses. Extensive experiments show that our model achieves state-of-the-art performance on two large-scale public datasets.